Weighted
least square
(Reading:
Faraway(2005, 1st edition), 6.2)
Look at
here
for a description of the data
that we will use in this section.
> p <- read.table("phy.data", header=T)
# note the extra argument to cope with not having row labels
> p # take a look of the data
momentum
energy crossx sd
1 4
0.345 367 17
2 6
0.287 311 9
3 8
0.251 295 9
...deleted...
10 150
0.060 192 5
Notice that
-
The sd decreases along with the reduction of energy.
-
Since the weights should be inversely proportional
to the variance, this suggests that the weights should be set to 1/sd2.
> x <- p[, 2]
# extract the energy variable, the predictor
> y <- p[, 3]
# extract the crossx variable,
the response
> w <- 1/p[, 4]^2
# the weights
> g <- lm(y~x, weights=w)
# fit the model with w as weights. "weights"
adds as an option.
> summary(g)
# take a look of the results
Call:
lm(formula = y ~
x, weights = w)
Residuals:
Min 1Q Median 3Q Max
-2.323e+00
-8.842e-01 1.266e-06 1.390e+00 2.335e+00
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept)
148.473 8.079 18.38 7.91e-08 ***
x
530.835 47.550 11.16 3.71e-06 ***
---
Signif. codes: 0
'***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard
error: 1.657 on 8 degrees of freedom
Multiple
R-Squared: 0.9397, Adjusted R-squared: 0.9321
F-statistic:
124.6 on 1 and 8 DF, p-value: 3.710e-06
-
We can test the significance of x by looking at the p-value from the
coefficients table or by looking at the overall F-stat (they are equivalent,
11.162=124.6, because this is a simple regression case).
-
But, if we want to do it the hard way, we must construct
the analysis of variance table by hand as follows:
> sum(w*g$res*g$res)
# the residual sum of squares
> wm <- sum(w*y)/sum(w)
# the weighted mean of y
> sum(w*(y-wm)*(y-wm))
# the weighted total SS
> 363.944 - 21.95265
# the regression SS
> (341.9914/1)/(21.95265/8)
# the overall F statistic
Let's try
fitting the regression without weights and see what the difference is:
> g1 <- lm(y~x)
# fit the model without
weights
> summary(g1)
# take a look of the fitted model
Call:
lm(formula = y ~
x)
Residuals:
Min 1Q
Median 3Q Max
-14.773 -9.319
-2.829 5.571 19.818
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept)
135.00 10.08 13.40 9.21e-07 ***
x
619.71 47.68 13.00 1.16e-06 ***
---
Signif. codes: 0
'***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard
error: 12.69 on 8 degrees of freedom
Multiple
R-Squared: 0.9548, Adjusted R-squared: 0.9491
F-statistic:
168.9 on 1 and 8 DF, p-value: 1.165e-06
-
Compare the output to the
summary of model g. Explain their differences.
-
R-square: why would g1 has a higher value than
g?
-
σ-hat: Why are
they so different? Do they estimate the same parameter?
-
residuals: Why are their ranges so different?
-
The
two fits can be compared:
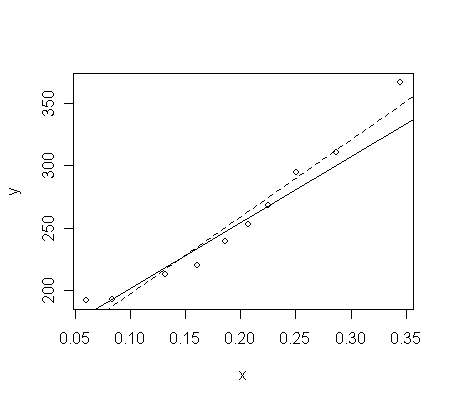
> plot(x,y)
# scatter plot of x and y
> abline(g)
# the fitted line of model g,
solid line
> abline(g1, lty=2)
# the fitted line of model
g2, dashed line
-
The un-weighted fit (dashed line) appears to fit the data better overall
-
But remember that for
lower values of energy, the variance in the response is less.
-
The weighted
fit (solid line) tries to catch these points with lower energy better than the others.
Weighted
least square can be regarded as regressing sqrt(wi)xi on
sqrt(wi)yi using ordinary least square:
> ym<-sqrt(w)*y
# calculate sqrt(wi)yi
> xm<-sqrt(w)*x
# calculate sqrt(wi)xi
>
g2 <- lm(ym~sqrt(w)+xm-1)
# regressing sqrt(wi)xi on
sqrt(wi)yi. "-1" drop the intercept.
>
summary(g2) # take a
look of the results
Call:
lm(formula = ym ~
sqrt(w) + xm - 1)
Residuals:
Min 1Q Median 3Q Max
-2.323e+00
-8.842e-01 1.266e-06 1.390e+00 2.335e+00
Coefficients:
Estimate
Std. Error t value Pr(>|t|)
sqrt(w)
148.473 8.079 18.38 7.91e-08 ***
xm
530.835 47.550 11.16 3.71e-06 ***
---
Signif. codes: 0
'***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard
error: 1.657 on 8 degrees of freedom
Multiple
R-Squared: 0.9983, Adjusted R-squared: 0.9978
F-statistic:
2303 on 2 and 8 DF, p-value: 9.034e-12
-
Compare the output to the summary of model g.
What quantities are different?
Explain why.
-
To get the correct overall F-test, we can use:
> anova(lm(ym~sqrt(w)-1), g2)
# the correct F-statistics
Analysis of
Variance Table
Model 1: ym ~
sqrt(w) - 1
Model 2: ym ~
sqrt(w) + xm - 1
Res.Df RSS
Df Sum of Sq F Pr(>F)
1 9
363.94
2 8 21.95
1 341.99 124.63 3.710e-06 ***
---
Signif. codes: 0
'***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Let's also check their residuals.
> print(g$res, digits=2)
# the residuals of model g
1
2 3 4 5 6 7 8 9 10
35.389 10.177
13.287 0.089 -5.356 -8.209 -13.938 -5.544 -0.063 11.677
> print(g2$res, digits=2)
# the residuals of model g2
1
2 3 4 5 6 7 8 9 10
2.082 1.131
1.476 0.013 -0.765 -1.368 -2.323 -0.924 -0.013 2.335
-
Why are the two groups of
residuals so
different?
-
Which residuals is yi
minus yi-hat?
Which residuals is
sqrt(wi)yi minus sqrt(wi)yi-hat?
-
Which residuals can be used to
perform diagnostics (i.e., residual analysis) and to calculate σ-hat by
directly taking sum of their squares?
-
Let's find out the relationship between the
two groups of residuals:
> print(sqrt(w)*g$res, digits=2)
# check if it equals residuals of model g2
1
2 3 4 5 6 7 8 9 10
2.082 1.131
1.476 0.013 -0.765 -1.368 -2.323 -0.924 -0.013 2.335