Generalized
least square
(Reading:
Faraway(2005, 1st edition), 6.1)
We'll
use a built-in R dataset called Longley's regression data where
-
the response is
number of people employed, yearly from 1947 to 1962, and
-
the predictors are
-
GNP
implicit price deflator (1954=100),
-
GNP,
-
unemployed,
-
armed forces,
-
non-institutionalized population 14 years of
age and over, and
-
year.
-
The Longley data was collected over 16 years and it is reasonable to expect that
there might be some correlation between errors in successive years.
Let us take a look of some background knowledge of the
data set.
>
help(longley)
Longley's Economic Regression Data
Description:
A macroeconomic data set which provides a well-known example for a highly
collinear regression.
Usage:
longley
Format:
A data frame with 7 economical variables, observed yearly from 1947 to 1962
(n=16).
GNP.deflator: GNP implicit price deflator (1954=100)
GNP: Gross National Product.
Unemployed: number of unemployed.
Armed.Forces: number of people in the armed forces.
Population: 'noninstitutionalized' population >= 14 years of age.
Year: the year (time).
Employed: number of people employed.
... deleted ...
> data(longley)
# load the data into R
> longley
# take a look of the data
GNP.deflator
GNP Unemployed Armed.Forces Population Year Employed
1947 83.0
234.289 235.6 159.0 107.608 1947 60.323
1948 88.5
259.426 232.5 145.6 108.632 1948 61.122
1949 88.2
258.054 368.2 161.6 109.773 1949 60.171
...deleted...
1962 116.9
554.894 400.7 282.7 130.081 1962 70.551
> emp <- longley[,7]
# extract the Employed variable
> gnp <- longley[,2]
# extract the GNP variable
> pop <- longley[,5]
# extract the Population variable
Let's fit a model with
> g <- lm(emp~gnp+pop)
# fit the model
> summary(g,cor=T)
# summary
of the fitted model
Call:
lm(formula = emp
~ gnp + pop)
Residuals:
Min
1Q Median 3Q Max
-0.80899 -0.33282
-0.02329 0.25895 1.08800
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept)
88.93880 13.78503 6.452 2.16e-05 ***
gnp
0.06317 0.01065 5.933 4.96e-05 ***
pop
-0.40974 0.15214 -2.693 0.0184 *
---
Signif. codes: 0
'***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard
error: 0.5459 on 13 degrees of freedom
Multiple
R-Squared: 0.9791, Adjusted R-squared: 0.9758
F-statistic:
303.9 on 2 and 13 DF, p-value: 1.221e-11
Correlation of
Coefficients:
(Intercept)
gnp
gnp
0.98
pop -1.00
-0.99
Note that
-
the model is built under the assumption that
the errors are uncorrelated.
-
Check the correlation between the estimates of
predictors gnp and pop. What do you notice?
> cor(gnp, pop)
# correlation between two predictors
-
Compare it to the correlation of coefficient. Note that
one is positive and another is negative. Explain why.
-
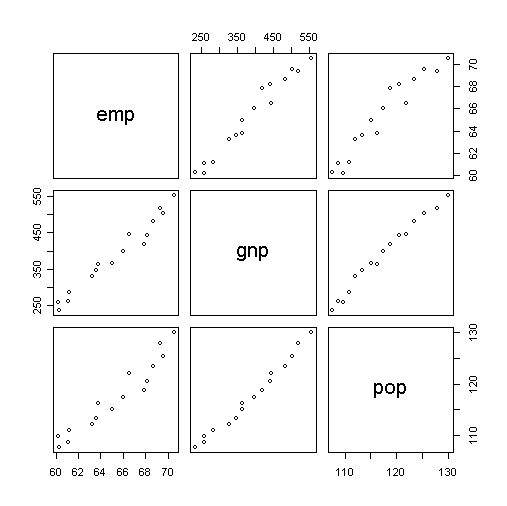
Now, let's take a look of their scatter plots.
> pairs(cbind(emp,gnp,pop)) # draw pairwise
scatter plots
What have you found from the plots?
Notice that
We will
calculate the results with correlated errors "by hand" below and compare them to
those assuming uncorrelated errors.
> x <- cbind(rep(1,16),gnp,pop)
# extract the X-matrix
> Sig <- diag(16)
# start to construct the
Σ matrix
> Sig
# diagonal entries in the correlation matrix are always one
> Sig[abs(row(Sig)-col(Sig))==1] <- 0.5
# assign the off-diagonal values of
Σ
> Sig # the
Σ matrix as we want it
[,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8] [,9] [,10] [,11] [,12] [,13] [,14]
[,15] [,16]
[1,] 1.0 0.5 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0
0.0 0.0 0.0
[2,] 0.5 1.0 0.5 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0
0.0 0.0 0.0
[3,] 0.0 0.5 1.0 0.5 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0
0.0 0.0 0.0
[4,] 0.0 0.0 0.5 1.0 0.5 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0
0.0 0.0 0.0
[5,] 0.0 0.0 0.0 0.5 1.0 0.5 0.0 0.0 0.0 0.0 0.0 0.0 0.0
0.0 0.0 0.0
[6,] 0.0 0.0 0.0 0.0 0.5 1.0 0.5 0.0 0.0 0.0 0.0 0.0 0.0
0.0 0.0 0.0
[7,] 0.0 0.0 0.0 0.0 0.0 0.5 1.0 0.5 0.0 0.0 0.0 0.0 0.0
0.0 0.0 0.0
[8,] 0.0 0.0 0.0 0.0 0.0 0.0 0.5 1.0 0.5 0.0 0.0 0.0 0.0
0.0 0.0 0.0
[9,] 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.5 1.0 0.5 0.0 0.0 0.0
0.0 0.0 0.0
[10,] 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.5 1.0 0.5 0.0 0.0
0.0 0.0 0.0
[11,] 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.5 1.0 0.5 0.0
0.0 0.0 0.0
[12,] 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.5 1.0 0.5
0.0 0.0 0.0
[13,] 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.5 1.0
0.5 0.0 0.0
[14,] 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.5
1.0 0.5 0.0
[15,] 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0
0.5 1.0 0.5
[16,] 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0
0.0 0.5 1.0
> Sigi <- solve(Sig)
# the Σ-1
> xtxi <- solve(t(x) %*% Sigi %*% x)
# calculate(XTΣ-1X)-1
> beta <- xtxi %*% t(x) %*% Sigi %*% emp
# β-hat = (XTΣ-1X)-1XTΣ-1Y
> beta # the GLS
estimate of
β
[,1]
113.93620168
gnp
0.08185425
pop
-0.68390315
> res <- emp-(x%*%beta)
# the residuals for this fit
> sig <- sqrt((t(res) %*% Sigi %*% res)/g$df)
# σ-hat is
estimated
> sig
# the value of
σ-hat
> sqrt(diag(xtxi))*sig
# s.e.(β-hati)=sqrt((XTΣ-1X)-1ii)*(σ-hat), the standard errors for
β-hat allowing for the correlation
[1]
9.760394461 0.007201431 0.105483848
The simplest way to model the correlation between
successive errors is the autoregressive (AR) form:
εt+1
= ρεt + δt,
where δt~N(0,τ2).
A rough estimate of the correlation ρ
is:
> cor(g$res[-1], g$res[-16])
# calculate cor(εt,
εt+1)
Under the AR(1) assumption,
We now construct the Σ
matrix and compute the GLS estimate of β along
with its standard errors.
> Sigma <- diag(16)
# start to calculate
Σ
> Sigma <- 0.31041^abs(row(Sigma)-col(Sigma))
# assign the off-diagonal values of
Σ
> print(Sigma, digits=1)
# take a look of
Σ
[,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8] [,9] [,10]
[,11] [,12] [,13] [,14] [,15] [,16]
[1,] 1e+00 3e-01 1e-01 3e-02 9e-03 3e-03 9e-04 3e-04 9e-05 3e-05
8e-06 3e-06 8e-07 2e-07 8e-08 2e-08
[2,] 3e-01 1e+00 3e-01 1e-01 3e-02 9e-03 3e-03 9e-04 3e-04 9e-05
3e-05 8e-06 3e-06 8e-07 2e-07 8e-08
[3,] 1e-01 3e-01 1e+00 3e-01 1e-01 3e-02 9e-03 3e-03 9e-04 3e-04
9e-05 3e-05 8e-06 3e-06 8e-07 2e-07
[4,] 3e-02 1e-01 3e-01 1e+00 3e-01 1e-01 3e-02 9e-03 3e-03 9e-04
3e-04 9e-05 3e-05 8e-06 3e-06 8e-07
[5,] 9e-03 3e-02 1e-01 3e-01 1e+00 3e-01 1e-01 3e-02 9e-03 3e-03
9e-04 3e-04 9e-05 3e-05 8e-06 3e-06
[6,] 3e-03 9e-03 3e-02 1e-01 3e-01 1e+00 3e-01 1e-01 3e-02 9e-03
3e-03 9e-04 3e-04 9e-05 3e-05 8e-06
[7,] 9e-04 3e-03 9e-03 3e-02 1e-01 3e-01 1e+00 3e-01 1e-01 3e-02
9e-03 3e-03 9e-04 3e-04 9e-05 3e-05
[8,] 3e-04 9e-04 3e-03 9e-03 3e-02 1e-01 3e-01 1e+00 3e-01 1e-01
3e-02 9e-03 3e-03 9e-04 3e-04 9e-05
[9,] 9e-05 3e-04 9e-04 3e-03 9e-03 3e-02 1e-01 3e-01 1e+00 3e-01
1e-01 3e-02 9e-03 3e-03 9e-04 3e-04
[10,] 3e-05 9e-05 3e-04 9e-04 3e-03 9e-03 3e-02 1e-01 3e-01 1e+00
3e-01 1e-01 3e-02 9e-03 3e-03 9e-04
[11,] 8e-06 3e-05 9e-05 3e-04 9e-04 3e-03 9e-03 3e-02 1e-01 3e-01
1e+00 3e-01 1e-01 3e-02 9e-03 3e-03
[12,] 3e-06 8e-06 3e-05 9e-05 3e-04 9e-04 3e-03 9e-03 3e-02 1e-01
3e-01 1e+00 3e-01 1e-01 3e-02 9e-03
[13,] 8e-07 3e-06 8e-06 3e-05 9e-05 3e-04 9e-04 3e-03 9e-03 3e-02
1e-01 3e-01 1e+00 3e-01 1e-01 3e-02
[14,] 2e-07 8e-07 3e-06 8e-06 3e-05 9e-05 3e-04 9e-04 3e-03 9e-03
3e-02 1e-01 3e-01 1e+00 3e-01 1e-01
[15,] 8e-08 2e-07 8e-07 3e-06 8e-06 3e-05 9e-05 3e-04 9e-04 3e-03
9e-03 3e-02 1e-01 3e-01 1e+00 3e-01
[16,] 2e-08 8e-08 2e-07 8e-07 3e-06 8e-06 3e-05 9e-05 3e-04 9e-04
3e-03 9e-03 3e-02 1e-01 3e-01 1e+00
We will try another
way, regressing S-1y on S-1x, to get the results:
> sm <- chol(Sigma)
# calculate ST, where
Σ=SST.
the "chol" command compute the Choleski factorization
> smi <- solve(t(sm))
# calculate S-1
> sx <- smi %*% x
# calculate S-1x
> sy <- smi %*% emp
# calculate S-1y
> g2 <- lm(sy~sx-1)
# regress S-1y on S-1x,
remember to exclude the constant term
> summary(g2, cor=T)
# take a look of the results
Call:
lm(formula =
sy ~ sx - 1)
Residuals:
Min
1Q Median 3Q Max
-0.74460
-0.34013 -0.02145 0.27667 1.07243
Coefficients:
Estimate
Std. Error t value Pr(>|t|)
sx
94.89889 13.94477 6.805 1.25e-05 ***
sxgnp
0.06739 0.01070 6.296 2.76e-05 ***
sxpop
-0.47427 0.15339 -3.092 0.00858 **
---
Signif.
codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual
standard error: 0.5424 on 13 degrees of freedom
Multiple
R-Squared: 0.9999, Adjusted R-squared: 0.9999
F-statistic:
4.312e+04 on 3 and 13 DF, p-value: < 2.2e-16
Correlation of
Coefficients:
sx
sxgnp
sxgnp
0.98
sxpop -1.00
-0.99
In practice,
Our initial estimate is 0.31041, but
once we fit our GLS model we'd need to re-estimate ρ
as:
> cor((t(sm)%*%g2$res)[-1], (t(sm)%*%g2$res)[-16])
# result using correct residuals, i.e., S*(residual)
-
Note that you cannot do this using the residuals of g2
(Why?). Check it out:
> cor(g2$res[-1], g2$res[-16])
# result using wrong
residuals
And
then,
This is cumbersome.
A more convenient approach may be found in the "nlme"
package, which contains a GLS fitting function. The approach is to
We will use the approach to fit
the model below (remember to add the "nlme" package if you haven't done it):
> library(nlme)
# load the "nlme" package.
> help(gls)
# take a look of the document of "gls" command.
Pay some
attention to the "correlation" option.
> year <- longley[,6]
# extract the Year variable
Let us fit the model with correlated errors
following an AR(1) structure.
> g3 <- gls(emp~gnp+pop, correlation=corAR1(form=~year))
# Use "help(corClasses)" and
"help(corAR1)" to get more information about correlation structure.
> summary(g3)
# take a look of the fitted
model
Generalized
least squares fit by REML
Model: emp ~
gnp + pop
Data: NULL
AIC BIC logLik
44.66377
47.48852 -17.33188
Correlation
Structure: AR(1)
Formula:
~year
Parameter
estimate(s):
Phi
0.6441692
Coefficients:
Value Std.Error t-value p-value
(Intercept)
101.85813 14.198932 7.173647 0.0000
gnp
0.07207 0.010606 6.795485 0.0000
pop
-0.54851 0.154130 -3.558778 0.0035
Correlation:
(Intr) gnp
gnp 0.943
We
can see that
-
the estimated value of ρ
is 0.6441692
-
however, if we check the
confidence intervals for ρ:
> intervals(g3)
# calculate confidence intervals of parameters
Approximate
95% confidence intervals
Coefficients:
lower est. upper
(Intercept)
71.18320461 101.85813306 132.5330615
gnp
0.04915865 0.07207088 0.0949831
pop
-0.88149053 -0.54851350 -0.2155365
attr(,"label")
[1]
"Coefficients:"
Correlation
structure:
lower est. upper
Phi -0.4444668
0.6441692 0.9646106
attr(,"label")
[1]
"Correlation structure:"
Residual
standard error:
lower
est. upper
0.2474787
0.6892070 1.9193820