Hierarchical or Nested Multinomial Responses (Reading: Faraway (2006), section 5.2)
Consider the data collected by Lowe et al. (1971) concerning live births with deformations of the central nervous system in south Wales. Let us first read the data into R and take a look:
>
cns <- read.table("cns.txt")
>
cns
Area NoCNS An Sp Other Water Work
1 Cardiff 4091 5 9 5 110 NonManual
2 Newport 1515 1 7 0 100 NonManual
3 Swansea 2394 9 5 0 95 NonManual
4 GlamorganE 3163 9 14 3 42 NonManual
5 GlamorganW 1979 5 10 1 39 NonManual
6 GlamorganC 4838 11 12 2 161 NonManual
7 MonmouthV 2362 6 8 4 83 NonManual
8 MonmouthOther 1604 3 6 0 122 NonManual
9 Cardiff 9424 31 33 14 110 Manual
10 Newport 4610 3 15 6 100 Manual
11 Swansea 5526 19 30 4 95 Manual
12 GlamorganE 13217 55 71 19 42 Manual
13 GlamorganW 8195 30 44 10 39 Manual
14 GlamorganC 7803 25 28 12 161 Manual
15 MonmouthV 9962 36 37 13 83 Manual
16 MonmouthOther 3172 8 13 3 122 Manual
In the dataset,
the variable NoCNS indicates no central nervous system (CNS) malformation
the variable An denotes anencephalus
the variable Sp denotes spina bifida
the variable Other represents other malformations
the variable Water is water hardness
subjects are categorized by the type of work performed by the parents (the variable Work)
Notice that the Area is confounded with the Water
¡@
We might consider a multinomial response with the four categories: NoCNS, An, Sp, and Other. However,
we can see that most births suffer no malformation and so this category dominate the other three
it is better to consider this as a hierarchical response as in the lecture note
We now separately develop a binomial model for whether malformation occurs and a multinomial model for the type of malformation.
¡@
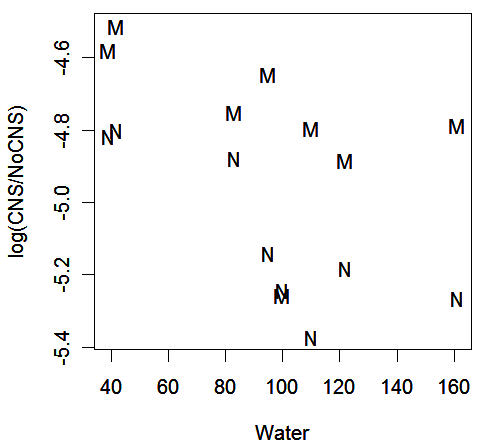
We start with the binomial model. First, we accumulate the number of CNS births and plot the data with the response on the logit scale:
>
cns$CNS <- cns$An+cns$Sp+cns$Other
>
plot(log(CNS/NoCNS) ~ Water, cns, pch=as.character(Work))

We observe that:
the proportion of CNS births falls with increasing water hardness
the proportion of CNS births is higher for manual workers
one observation (Manual, Newport, Water=100, logit=-5.26) may be an outlier
¡@
Because Area is confounded with Water, we cannot put both these predictors in one model. Let us try them both in different models and compare:
> binmodw <- glm(cbind(CNS,NoCNS) ~ Water + Work, cns, family=binomial)
>
binmoda <- glm(cbind(CNS,NoCNS) ~ Area + Work, cns, family=binomial)
>
anova(binmodw,binmoda,test="Chi")
Analysis of Deviance Table
Model 1: cbind(CNS, NoCNS) ~ Water + Work
Model 2: cbind(CNS, NoCNS) ~ Area + Work
Resid. Df Resid. Dev Df Deviance P(>|Chi|)
1 13 12.3628
2 7 3.0771 6 9.2857 0.1581
Notice that:
if Water is regarded as a nominal variable, it will has the same effect as Area
therefore, one can view this test as a check for linear trend in the effect of water hardness
we find that the simpler model using Water is acceptable
a check for an interaction effect revealed nothing significant (exercise)
a look at the the residuals is worthwhile:
> halfnorm(residuals(binmodw))
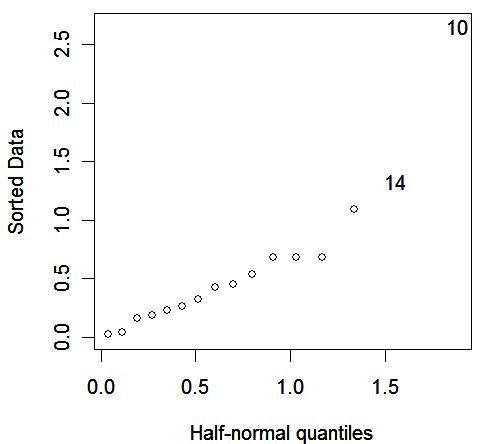
We see an outlier corresponding to Newport manual workers. The case deserves closer examination.
> summary(binmodw)
Call:
glm(formula = cbind(CNS, NoCNS) ~ Water + Work, family = binomial,
data = cns)
Deviance Residuals:
Min 1Q Median 3Q Max
-2.65570 -0.30179 -0.03131 0.57213 1.32998
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -4.4325803 0.0897889 -49.367 < 2e-16 ***
Water -0.0032644 0.0009684 -3.371 0.000749 ***
WorkNonManual -0.3390577 0.0970943 -3.492 0.000479 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 41.047 on 15 degrees of freedom
Residual deviance: 12.363 on 13 degrees of freedom
AIC: 102.49
Number of Fisher Scoring iterations: 4
We see that:
the residual deviance is close to the degrees of freedom indicating a reasonable fit to the data
since:
> exp(-0.3390577)
[1] 0.7124413
births to non-manual workers have a 29% lower chance of CNS malformation
water hardness ranges from about 40 to 160. So, a difference of 120 would decrease the odds of CNS malformation by about 32%:
> exp(120*(-0.0032644))
[1] 0.6758879
Now, consider a multinomial model for the three
malformation types conditional on a malformation having occurred. As this data
is grouped, it is most convenient to present the response as a matrix formed by
An, Sp,
and Other:
> library(nnet)
> cmmod <- multinom(cbind(An,Sp,Other) ~ Water + Work, cns)
# weights: 12 (6 variable)
initial value 762.436928
iter 10 value 685.762336
final value 685.762238
converged
¡@
Let us use the AIC criterion to select important effects:
> nmod <- step(cmmod)
Start: AIC= 1383.52
cbind(An, Sp, Other) ~ Water + Work
trying - Water
# weights: 9 (4 variable)
initial value 762.436928
iter 10 value 686.562074
final value 686.562063
converged
trying - Work
# weights: 9 (4 variable)
initial value 762.436928
final value 686.580556
converged
Df AIC
- Water 4 1381.124
- Work 4 1381.161
<none> 6 1383.524
# weights: 9 (4 variable)
initial value 762.436928
iter 10 value 686.562074
final value 686.562063
converged
Step: AIC= 1381.12
cbind(An, Sp, Other) ~ Work
trying - Work
# weights: 6 (2 variable)
initial value 762.436928
final value 687.227416
converged
Df AIC
- Work 2 1378.455
<none> 4 1381.124
# weights: 6 (2 variable)
initial value 762.436928
final value 687.227416
converged
Step: AIC= 1378.45
cbind(An, Sp, Other) ~ 1
We see that:
the model selection procedure ends up with a model containing only the intercept, i.e., neither predictor has significant effect.
the fitted model is:
> summary(nmod, cor=F)
Call:
multinom(formula = cbind(An, Sp, Other) ~ 1, data = cns)
Coefficients:
(Intercept)
Sp 0.2896333
Other -0.9808293
Std. Errors:
(Intercept)
Sp 0.08264518
Other 0.11967839
Residual Deviance: 1374.455
AIC: 1378.455
the fitted proportions are:
>
cc <- c(0, 0.2896333, -0.9808293)
>
names(cc) <- c("An","Sp","Other")
>
exp(cc)/sum(exp(cc))
An Sp Other
0.3688761 0.4927954 0.1383285
¡@
So, we find that
water hardness and parents' profession are related to the probability of a malformed birth
but, they have no effect on the type of malformation
if we fit a multinomial logit model to all 4 categories and use NoCNS as the baseline category:
> amod <- multinom(cbind(NoCNS,An,Sp,Other) ~ Water + Work, cns)
# weights: 16 (9 variable)
initial value 117209.801938
iter 10 value 27783.394982
iter 20 value 20770.391437
iter 30 value 11403.858565
iter 40 value 5239.204601
iter 50 value 4716.675652
final value 4695.482597
converged
Call:
multinom(formula = cbind(NoCNS, An, Sp, Other) ~ Water + Work,
data = cns)
Coefficients:
(Intercept) Water WorkNonManual
An -5.455141 -0.002908840 -0.3638797
Sp -5.071047 -0.004323052 -0.2435884
Other -6.594670 -0.000513569 -0.6421775
Residual Deviance: 9390.965
AIC: 9408.965
We find that
both Water and Work are significant (exercise)
the fact that Water and Work do not distinguish the type of malformation is not easily discovered from this model
¡@
¡@
¡@
¡@
¡@