Negative Binomial Response (Reading: Faraway (2006), section 3.3)
For a count response yx with the property Var(yx)>>E(yx), an alternative to the dispersion-parameter approach is to model yx as negative-binomially distributed, where the negative binomial records the number of "failures" before the rth "success" in a series of i.i.d. Bernoulli trials (rather than the total number of trials before the rth success). We will demonstrate the negative-binomial approach using a data from an experiment performed by ATT. The experiment varied five factors relevant to a wave-soldering procedure for mounting components on printed circuit boards. The response variable, skips, is a count of how many solder skips appeared to a visual inspection. The data comes from Comizzoli, Lanwehr, and Sinclair (1990). Let us first read the data into R and take a look at it:
> solder <- read.table("solder.txt")
> solder
Opening Solder Mask PadType Panel skips
1 L Thick A1.5 W4 1 0
2 L Thick A1.5 W4 2 0
3 L Thick A1.5 W4 3 0
ˇKdeletedˇK
900 S Thin B6 L9 3 15
ˇ@
Let us start the analysis by fitting an ordinary Poisson GLM (i.e., s2=1) to the data:
>
modp <- glm(skips ~ . , family=poisson, data=solder)
> summary(modp)
Call:
glm(formula = skips ~ ., family = poisson, data = solder)
Deviance Residuals:
Min 1Q Median 3Q Max
-3.6413 -1.2562 -0.5286 0.5670 4.7289
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -1.10042 0.09267 -11.874 < 2e-16 ***
OpeningM 0.57161 0.05707 10.017 < 2e-16 ***
OpeningS 1.81475 0.05044 35.980 < 2e-16 ***
SolderThin 0.84682 0.03327 25.453 < 2e-16 ***
MaskA3 0.51315 0.07098 7.230 4.83e-13 ***
MaskA6 1.81103 0.06609 27.404 < 2e-16 ***
MaskB3 1.20225 0.06697 17.953 < 2e-16 ***
MaskB6 1.86648 0.06310 29.580 < 2e-16 ***
PadTypeD6 -0.40328 0.06028 -6.690 2.24e-11 ***
PadTypeD7 -0.08979 0.05521 -1.626 0.103852
PadTypeL4 0.22460 0.05117 4.389 1.14e-05 ***
PadTypeL6 -0.70633 0.06637 -10.642 < 2e-16 ***
PadTypeL7 -0.48260 0.06176 -7.814 5.52e-15 ***
PadTypeL8 -0.30778 0.05862 -5.251 1.51e-07 ***
PadTypeL9 -0.63793 0.06489 -9.831 < 2e-16 ***
PadTypeW4 -0.19021 0.05671 -3.354 0.000796 ***
PadTypeW9 -1.56252 0.09165 -17.048 < 2e-16 ***
Panel 0.14670 0.01745 8.405 < 2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for poisson family taken to be 1)
Null deviance: 8788.2 on 899 degrees of freedom
Residual deviance: 1829.0 on 882 degrees of freedom
AIC: 3967.6
Number of Fisher Scoring iterations: 5
Notice that:
all five factors are significant in this analysis, but
the model has a residual deviance of 1829 on 882 degrees of freedom, which suggests
this is not a good fit
the significances of the effects may not be reliable because the standard errors of the estimates of the effects are under-estimated if overdispersion exists
Let us examine whether we can change the
Xb
structure by including all 2-factor interactions to improve the deviance to an
acceptable level:
>
modp2 <- glm(skips ~ (Opening +Solder + Mask + PadType + Panel)^2 , family=poisson,
data=solder)
> anova(modp,modp2,test="Chi")
Analysis of Deviance Table
Model 1: skips ~ Opening + Solder + Mask + PadType + Panel
Model 2: skips ~ (Opening + Solder + Mask + PadType + Panel)^2
Resid. Df Resid. Dev Df Deviance P(>|Chi|)
1 882 1829.00
2 790 1068.82 92 760.19 9.99e-106
The difference-in-deviance test has a very small p-value=9.99e-106, which shows that the model with interactions is a much better fit than the one without interactions. However, let us check whether the new model can pass the goodness-of-fit test:
> deviance(modp2)
[1] 1068.817
> df.residual(modp2)
[1] 790
> pchisq(1068.817,790,lower=FALSE)
[1] 1.130640e-10
Unfortunately,
it is not enough to conclude that the model fits
although we can try add more interactions (such as 3-factor interactions), it will make interpretation increasingly difficult
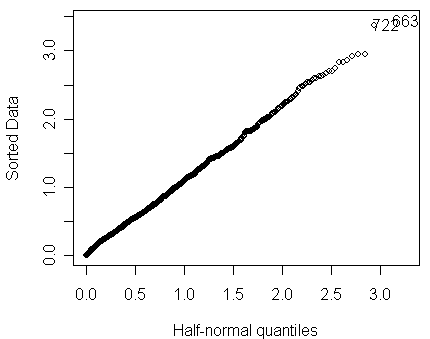
A check for outliers as shown below reveals no problem:
> halfnorm(residuals(modp2))

ˇ@
Suppose that overdispersion is the cause of large deviance. A negative binomial approach can be adopted. We can specify the value of r (a parameter of negative binomial). Notice that when r is fixed, the model is still an ordinary GLM. Here, we choose r=1 to demonstrate the method under a main-effect model although there is no substantive motivation from this application to use r=1:
>
library(MASS)
>
modn <- glm(skips ~ .,negative.binomial(1),solder)
> summary(modn)
Call:
glm(formula = skips ~ ., family = negative.binomial(1), data = solder)
Deviance Residuals:
Min 1Q Median 3Q Max
-1.9628 -0.8021 -0.2373 0.3204 2.1097
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -1.69933 0.16333 -10.404 < 2e-16 ***
OpeningM 0.50854 0.08797 5.781 1.03e-08 ***
OpeningS 1.99966 0.08069 24.783 < 2e-16 ***
SolderThin 1.04894 0.06364 16.483 < 2e-16 ***
MaskA3 0.65710 0.10463 6.281 5.29e-10 ***
MaskA6 2.52649 0.12123 20.841 < 2e-16 ***
MaskB3 1.27261 0.10779 11.806 < 2e-16 ***
MaskB6 2.08026 0.10482 19.845 < 2e-16 ***
PadTypeD6 -0.46118 0.13788 -3.345 0.000858 ***
PadTypeD7 0.01608 0.13350 0.120 0.904184
PadTypeL4 0.46883 0.13046 3.594 0.000344 ***
PadTypeL6 -0.47115 0.13799 -3.414 0.000668 ***
PadTypeL7 -0.29494 0.13620 -2.166 0.030616 *
PadTypeL8 -0.08493 0.13432 -0.632 0.527368
PadTypeL9 -0.52125 0.13854 -3.763 0.000179 ***
PadTypeW4 -0.14250 0.13481 -1.057 0.290776
PadTypeW9 -1.48361 0.15317 -9.686 < 2e-16 ***
Panel 0.16932 0.03811 4.443 1.00e-05 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for Negative Binomial(1) family taken to be 0.556042)
Null deviance: 1743.01 on 899 degrees of freedom
Residual deviance: 558.67 on 882 degrees of freedom
AIC: 3883.7
Number of Fisher Scoring iterations: 9
We could experiment with different values of r, but there is a more direct way of achieving this by allowing the parameter r to vary and be estimated using maximum likelihood (notice that when r is treated as a parameter, the model is not an ordinary GLM):
> library(MASS)
> modnr <- glm.nb(skips ~ .,solder)
> summary(modnr, cor=F)
Call:
glm.nb(formula = skips ~ ., data = solder, init.theta = 4.39715724541988,
link = log)
Deviance Residuals:
Min 1Q Median 3Q Max
-2.7376 -1.0068 -0.3834 0.4460 2.7829
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -1.42245 0.14274 -9.965 < 2e-16 ***
OpeningM 0.50294 0.07976 6.306 2.87e-10 ***
OpeningS 1.91317 0.07152 26.750 < 2e-16 ***
SolderThin 0.93932 0.05362 17.517 < 2e-16 ***
MaskA3 0.58981 0.09651 6.112 9.87e-10 ***
MaskA6 2.26734 0.10182 22.269 < 2e-16 ***
MaskB3 1.21101 0.09637 12.566 < 2e-16 ***
MaskB6 1.99037 0.09223 21.580 < 2e-16 ***
PadTypeD6 -0.46592 0.11238 -4.146 3.38e-05 ***
PadTypeD7 -0.03315 0.10673 -0.311 0.756114
PadTypeL4 0.38268 0.10265 3.728 0.000193 ***
PadTypeL6 -0.57844 0.11413 -5.068 4.01e-07 ***
PadTypeL7 -0.36656 0.11094 -3.304 0.000953 ***
PadTypeL8 -0.15890 0.10821 -1.468 0.141986
PadTypeL9 -0.56600 0.11393 -4.968 6.77e-07 ***
PadTypeW4 -0.20044 0.10873 -1.844 0.065255 .
PadTypeW9 -1.56460 0.13621 -11.486 < 2e-16 ***
Panel 0.16369 0.03139 5.214 1.85e-07 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for Negative Binomial(4.3972) family taken to be 1)
Null deviance: 4043.3 on 899 degrees of freedom
Residual deviance: 1008.3 on 882 degrees of freedom
AIC: 3683.3
Number of Fisher Scoring iterations: 1
Theta: 4.397
Std. Err.: 0.495
2 x log-likelihood: -3645.309
We can see that:
the estimate of r is 4.397 with a standard error of 0.495
when conditioning on r=4.397, we can use the usual inferential techniques for an ordinary GLM to compare the negative binomial model
we can compare the result with what obtained from the dispersion-parameter approach as given below:
> (dp <- sum(residuals(modp,type="pearson")^2)/modp$df.res)
[1] 2.022422
> summary(modp,dispersion=dp)
Call:
glm(formula = skips ~ ., family = poisson, data = solder)
Deviance Residuals:
Min 1Q Median 3Q Max
-3.6413 -1.2562 -0.5286 0.5670 4.7289
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -1.10042 0.13179 -8.350 < 2e-16 ***
OpeningM 0.57161 0.08115 7.044 1.87e-12 ***
OpeningS 1.81475 0.07173 25.301 < 2e-16 ***
SolderThin 0.84682 0.04731 17.898 < 2e-16 ***
MaskA3 0.51315 0.10094 5.084 3.70e-07 ***
MaskA6 1.81103 0.09398 19.270 < 2e-16 ***
MaskB3 1.20225 0.09523 12.624 < 2e-16 ***
MaskB6 1.86648 0.08974 20.800 < 2e-16 ***
PadTypeD6 -0.40328 0.08573 -4.704 2.55e-06 ***
PadTypeD7 -0.08979 0.07851 -1.144 0.252753
PadTypeL4 0.22460 0.07277 3.086 0.002026 **
PadTypeL6 -0.70633 0.09439 -7.483 7.26e-14 ***
PadTypeL7 -0.48260 0.08783 -5.495 3.91e-08 ***
PadTypeL8 -0.30778 0.08336 -3.692 0.000222 ***
PadTypeL9 -0.63793 0.09228 -6.913 4.74e-12 ***
PadTypeW4 -0.19021 0.08065 -2.358 0.018349 *
PadTypeW9 -1.56252 0.13034 -11.988 < 2e-16 ***
Panel 0.14670 0.02482 5.910 3.42e-09 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for poisson family taken to be 2.022422)
Null deviance: 8788.2 on 899 degrees of freedom
Residual deviance: 1829.0 on 882 degrees of freedom
AIC: 3967.6
Number of Fisher Scoring iterations: 5
Exercise: examine what is different and what is similar between the results from the negative-binomial and the dispersion-parameter approaches